

OmniPath is a database of molecular biology prior knowledge developed in Saez Lab and Korcsmaros Lab. It combines data from more than 100 resources and contains protein-protein and gene regulatory interactions, enzyme-PTM relationships, protein complexes, protein annotations and intercellular communication. Read more
More than ten workflows, tutorials and little examples. R, Python and web service queries. How to extract networks from OmniPath? How to build intercellular communication networks? How to combine OmniPath data with drug targets or tissue expression? Show me
With the OmnipathR package it's really easy to fetch data from the OmniPath web service into R data frames, build intercellular signaling networks and input data for NicheNet. Available from Bioconductor and github.
The Python client downloads OmniPath data into pandas data frames. It is also able to build intercellular (ligand-receptor) networks. Available from PyPI and github.
Load OmniPath networks directly into Cytoscape with the OmniPath plug-in. Read more about the plug-in in this paper.
The pypath Python module builds the OmniPath databases. It's super flexible, customizable, comes with versatile APIs and many useful utilities (e.g. ID translation). Available in PyPI and github.
Some questions we've already answered in the FAQ. Have a question about OmniPath data or tools? Something doesn't work? Need a new feature? Open an issue: pypath, OmniPath data, OmnipathR (R client), omnipath Python client, OmniPath Cytoscape plug-in. Pull requests are also welcome. Private messages: omnipathdb@gmail.com.
Examples of HTTP requests to the OmniPath web service.
Combined metabolic and signaling network in MetaPKN by Aurelien Dugourd.
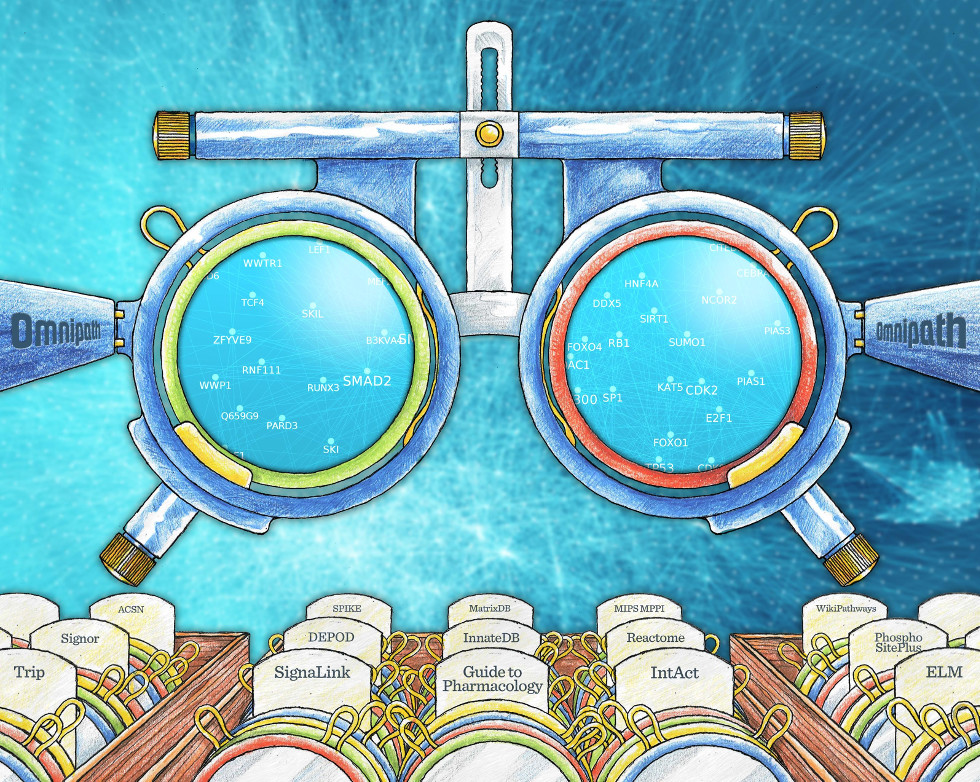
Illustration by Spencer Phillips, EMBL-EBI. This cover artwork is the result of a collaboration between Spencer Phillips, Mary Todd-Bergman, Denes Turei, Tamas Korcsmaros and Julio Saez-Rodriguez, starting with the initial idea of using lenses by Jakob Wirbel (RWTH-Aachen). A modified version of this figure appeared in the Cover of Nature Methods Dec 2016
OmniPath is a database of molecular biology prior knowledge. It combines data from over 100 resources and builds 5 integrated databases: signaling network (interactions), enzyme-PTM relationships, protein complexes, protein annotations (function, localization, tissue, disease, structure, etc), and intercellular communication roles (e.g. ligand, receptor; intercell). More information is available in our recent paper.
Overview of resources in pypath and OmniPath. New additions since Nov 2016 highlighted in violet. Expanded from Fig 1 of Turei et al. Nature Methods (2016). For high resolution PDF click here .
The OmniPath databases are built by the Python module pypath and are distributed by the web service at omnipathdb.org. The web service can be queried by any HTTP client, or the R/Bioconductor package OmnipathR and a Python client. These clients also provide some convenient methods for post-processing and are also available in Bioconductor and pypi. In addition, the R package provides an integration to NicheNet, a method for inferring ligand-receptor interaction and downstream pathways from scRNA data. Furthermore, the R package contains interface to 20 further resources which are independent from OmniPath.
OmniPath consists of five databases (top), each of them combine data from a number of original resources. This database knowledge can be used for the analysis of various omics data (middle) with various tools (bottom).
Database knowledge from OmniPath, together with transcriptomics data, can be used to infer signaling mechanisms such as transcription factor activities, pathway activities and ligand-receptor interaction activities. The names of some software tools are shown on the arrows.
Workflow and design: OmniPath is based on four major types of resources, and the pypath Python package combines the resources to build five databases. The databases are available by the database builder software pypath, the web resource at https://omnipathdb.org/, the R package OmnipathR, the Python client omnipath, the Cytoscape plug-in and can be exported to formats such as Biological Expression Language (BEL).
No, the original resources collect data from a variety of tissues, cell types and physiological conditions. In most of the cases this information is not even included with the data. Even if it would be included, we couldn't use it to obtain a, for example, T-cell specific signaling network, because the number of cell types and conditions is enormous. The same interactions present across many cell types and conditions, and at the same time, the signaling of T-cells also varies depending on the conditions and the sub-lineage. Instead of looking for cell type specific signaling networks, we recommend you to use omics data to contextualize the generic network from OmniPath. For example, use transcriptomics data to limit the network to proteins expressed in your cell type of interest. If you don't have such data, use public resources such as Expression Atlas, GEO, HumanBase, CCLE etc. An easy quick solution is to use the Human Protein Atlas which is also available in the OmniPath annotations database, we show this in the "Combining OmniPath annotations and networks" tutorial.
Is there a confidence score?The network, the enzyme-PTM relationships and complexes can be filtered by either the `curation_effort` value (unique resource-reference pairs) or by the number of references. By removing the records with no references one can easily select the literature curated part of the data. In the intercell database a `consensus_score` is available for each composite record: the number of resources annotating the protein for a certain category (e.g. if three resources agree that a protein is a transporter that’s more trustworthy compared to an annotation based on a single resource). We emphasize that these metrics express the likelihood that a piece of data is correct but has nothing to do with its biological significance: maybe a weak interaction is described by many resources while an important one by only one resource. Also such metrics are distorted by data integration across databases, for example the data collected orignally by PhosphoSite has been included in many other resources.
I am a company, can I use OmniPath?The data redistributed by the OmniPath web service doesn't have a license, each original resource carries over its license. You can use the license = "commercial" parameter in the web service queries and in the R and Python clients. This way you get only the records which allow commercial use. Here you find the license information of all the resources in OmniPath.
How often do you update OmniPath?We add new resources every couple of months. We also update all the resources at least once a year. Most of the original resources don't update their contents so often, and some of them do it at a higher frequency. Before every update we archive the data. A log of updates is also available here.
Some data in OmniPath seems to be wrongAll databases unavoidably contain some errors. The errors in OmniPath either come from the original resources or are introduced by a data processing mistake. We are very grateful if you report errors and we use the reports to improve the quality of OmniPath. If you found a particular wrong record, please report it by this form. Systematic errors affecting many records can be reported here. Also in this case one example of the wrong records is very helpful.
D Turei, A Valdeolivas, L Gul, N Palacio-Escat, M Klein, O Ivanova, M Olbei, A Gabor, F Theis, D Modos, T Korcsmaros and J Saez-Rodriguez (2021) Integrated intra- and intercellular signaling knowledge for multicellular omics analysis.
Mol Syst Biol
17: e9923
PMID: 33749993 |
PDF
F Ceccarelli, D Turei, A Gabor, J Saez-Rodriguez (2020) Bringing Data
From Curated Pathway Resources to Cytoscape With OmniPath.
Bioinformatics
36 (1)
PMID: 31886476
D Turei, T Korcsmaros and J Saez-Rodriguez (2016) OmniPath: guidelines
and gateway for literature-curated signaling pathway resources.
Nature Methods
13 (12)
PMID: 27898060 |
Full text |
Supplementary
Dénes Türei,
Alberto Valdeolivas,
Leila Gul,
Dezső Módos,
Francesco Ceccarelli,
Nicolàs Palacio,
Olga Ivanova,
Michal Klein,
Attila Gábor,
Márton Ölbei,
Saez Lab &
Korcsmaros Lab &
Theis Lab
2016-2022.
Feedback: omnipathdb@gmail.com